The protease network that regulates innate immunity in mosquitoes
The innate immune system of mosquitoes is a critical determinant of their vector competence. This includes the ability to support development and transmission of the protozoan parasite species in the genus Plasmodium by Anopheles mosquitoes, the principal vectors of human malaria worldwide. Insight into the regulation of innate immune effector mechanisms remains incomplete, but is vitally important to our fundamental understanding of host-pathogen interactions in this most important human vector-borne disease. The long-term goal is to understand immune system regulation in An. gambiae to inform current and future vector control strategies. The objective of this project is to globally identify mechanisms of immune system regulation by determining the interactions within the extracellular protease network that activate and link opsonization to melanization in the context of distinct microbial infections.
The NetSE group is contributing to the project by focusing on Aim 3. Under the third aim standard network science approaches are used to visualize all protease interactions in the system, and to analyze this network to infer proteolytic flow through that links opsonization and melanization and to identify the key molecules that control immunity. The proposed research is innovative, as it will for the first time evaluate protease cascades as a single, integrated network that controls mosquito humoral immunity during diverse immune challenges.
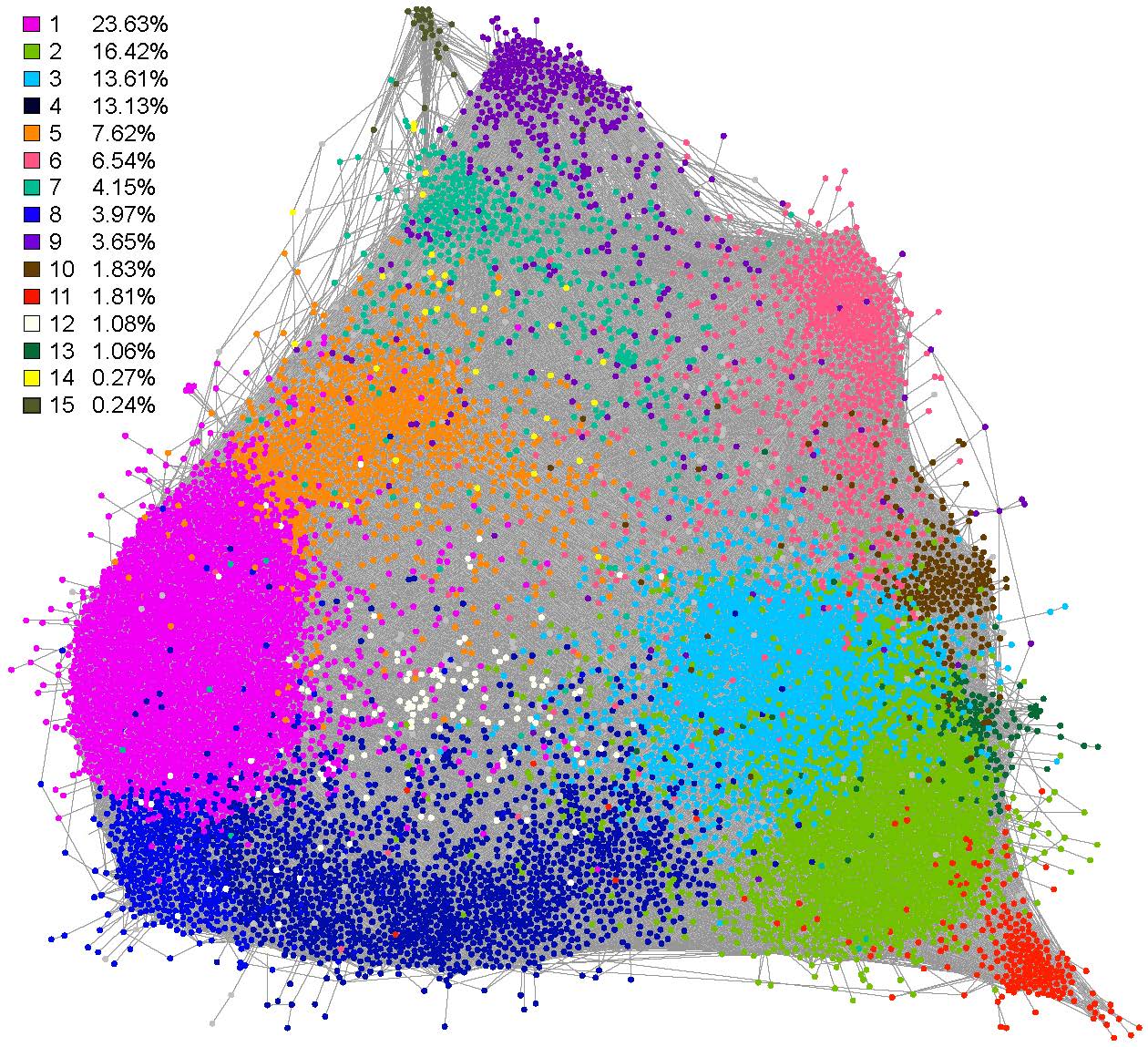 The gene expression network AgGCN1.0 consists of 11,794 nodes and 382,144 edges. The 15 major communities of the largest connected component in AgGCN1.0 are shown in the figure. The network was visualized with Gephi 0.9.2 using the ForceAtlas 2.0 algorithm. Communities are numbered consecutively based on the number of nodes they contain, and colored according to the panel on the left. The enrichment of GO terms and KEGG pathway analysis revealed that AgGCN1.0 is structured by gene function, maximizing integration of essential cellular processes and enabling evolutionary flexibility to integrate the expression of novel biological functions.
The gene expression network AgGCN1.0 consists of 11,794 nodes and 382,144 edges. The 15 major communities of the largest connected component in AgGCN1.0 are shown in the figure. The network was visualized with Gephi 0.9.2 using the ForceAtlas 2.0 algorithm. Communities are numbered consecutively based on the number of nodes they contain, and colored according to the panel on the left. The enrichment of GO terms and KEGG pathway analysis revealed that AgGCN1.0 is structured by gene function, maximizing integration of essential cellular processes and enabling evolutionary flexibility to integrate the expression of novel biological functions.
Duration September 1, 2018 - August 31, 2023
Investigators
Faculty
Kristin Michel (Google Profile) (PI)
Caterina Scoglio (Google Profile) (co-PI)
PhD Student
Products
Publications
Junyao Kuang, Caterina Scoglio
Layer reconstruction and missing link prediction of multilayer network with maximum a posteriori estimation
Physical Review E, vol.120, Issue 2, 2021
Junyao Kuang, Caterina Scoglio
A principled approach for weighted multilayer network aggregation
arXiv preprint arXiv:2103.05774, 2021
Junyao Kuang, Caterina Scoglio, Nicolas Buchon, Kristin Michel
Mining Anopheles gambiae gene co-expression across hundreds of experimental conditions with missing values
In preparation, 2021
Junyao Kuang, Caterina Scoglio
Network structure and feature learning from rich but noisy data
Submitted for journal publication, 2021
Data
AgGCN1.0
Supported by Agency National Institute of Health (NIH) Institute National Institute of Allergy and Infectious Diseases (NIAID) Type Research Project (R01). Project #1R01AI140760-01A12. Any opinions, findings, and conclusions or recommendations expressed in this website are those of the author(s) and do not necessarily reflect the views of the funding agency.